After PDI is installed, you can proceed with getting the sources for the hands-on tutorial from github.
Before compilation, configure the tutorial by detecting all dependencies:
Now you're ready to work, good luck!
For each exercise, once you've modified it, you can compile it by running the following command:
Where ? is the number of the exercise.
Then, you can run it with the following command:
Where ? is again the number of the exercise.
Ex1. implements a simple heat equation solver using an explicit forward finite difference scheme parallelized with MPI as described in the PDI example. If you want to better understand what's going on, you should read this description before continuing.
In the exercises however, PDI will only be used to decouple I/O operations. There is no need to fully dive in the core of the solver implemented in the iter and exchange functions. The specification tree in the .yml files and the main function are the locations where all the I/O-related aspects will be handled and the only ones you will actually need to fully understand or modify.
This example uses the paraconf library to read its parameters in yaml format from the ex1.yml file. If you've never heard about yaml, have a quick look at this example.
ex1.yml.ex1.yml to be able to run the code with 3 MPI processes.Ex2. is the same code as ex1. with PDI calls added in main function. The PDI Trace plugin is used to trace PDI calls.
PDI_share and PDI_reclaim calls to match the output of ex2.log file. Change only the ex2.c file. The calls for now don't need to make any logic, just match output to the ex2.log.Let's take the code from ex2. and make it output some HDF5 data. No need to touch the C code here, the PDI yaml file should be enough. The Trace plugin was replaced by the Dec'HDF5 plugin.
Fill 2 sections in the yaml file:
data section to indicate to PDI the type of the fields that are exposed.decl_hdf5 for the configuration of Dec'HDF5 plugin.Only dsize is written as of now, let's add psize and pcoord to match the content expected described in ex3.href 5dump (use h5dump command to see content of HDF5 file).
ex3.h5 file, because the data will not be overwritten.In this exercise each MPI process will write its local matrix to separete HDF5 files. Touch only the yaml file again.
This time write the real 2D data contained in main_field using 2 MPI processes.
Notice that a list to write multiple files was used in the decl_hdf5 section instead of a single mapping as before.
Unlike the other fields manipulated until now, the type of main_field is not fully known, its size is dynamic. By moving other fields in the metadata section, you can reference them from "$ expressions" in the configuration file. This can be used to specify a dynamic size for main_field.
Unlike the other fields manipulated until now, main_field is exposed multiple times. In order not to overwrite it every time it is exposed, you can add a when condition to restrict its output. Only write main_field at the second iteration (when ii==1).
Set the parallelism degree to 2 in height and try to match the expected content described in ex4.h5dump.
In ex4, there were 2 pieces of data to ex4-data*.h5, but the file is opened and closed for each and every write. Since Decl'HDF5 only sees the data appear one after the other, it does not keep the file open. Since ii and main_field are shared in an interlaced way, they are both available at the same time and could be written without opening the file twice. You have to use events for that.
There are 3 main tasks in this exercise:
loop when both ii and main_field are shared. With the Trace plugin, check that the event is indeed triggered at the expected time as described in ex5.log.on_event mechanism to trigger the write of ii and main_field. This mechanism can be combined with a when directive, in that case the write is only executed when both mechanisms agree.iter1 and iter2.Match the content as expected in ex5.h5dump.
As you can notice, the PDI code is quite redundant. In this exercise, you will use PDI_expose and PDI_multi_expose to simplify the code while keeping the exact same behaviour.
There are lots of matched PDI_share/PDI_reclaim in the code. Replace these by PDI_expose that is the exact equivalent of a PDI_share followed by a matching PDI_reclaim.
This replacement is not possible for interlaced PDI_share/PDI_reclaim with events in the middle. This case is however handled by PDI_multi_expose call that exposes all data, then triggers an event and finally does all the reclaim in reverse order. Replace the remaining PDI_share/PDI_reclaim by PDI_exposes and PDI_multi_exposes. There is one PDI_share/PDI_reclaim you can not replace.
Touch only the C file in this exercise.
Ensure that your code keeps the exact same behaviour by comparing its trace to ex6.log.
In this exercise, you will only write a selection (part) of the data to the HDF5 file.
As you can notice, now the dataset is independantly described in the file. Use two directives to specify a selection from the data to write and a selection in the dataset where to write.
memory_selection tells what part to take from the data.dataset_selection tells where to write this part of data in file.Restrict the selection to the second line from the data and write it to a one-dimensional dataset in file.
Touch only the yaml file in this exercise.
Match the expected output described in ex7.out.
Here is an example:
The graphical representation:
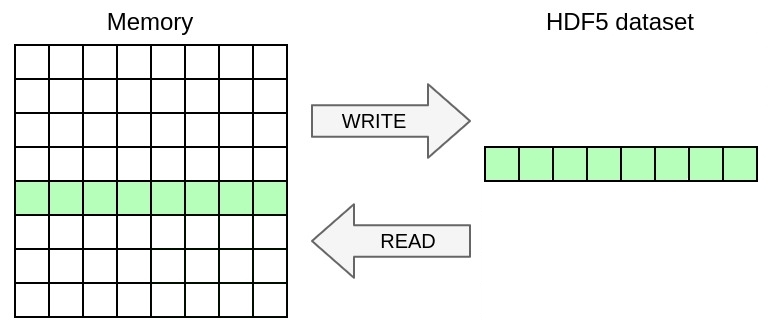
You can also add dimensions, write the 2D array excluding ghosts as a slab of a 3D dataset including a dimension for the time-iteration. Write iterations 1 to 3 inclusive into dimensions 0 to 2.
Touch only the yaml file in this exercise.
Match the expected output described in ex8.h5dump.
Here is an example:
And the graphical representation:
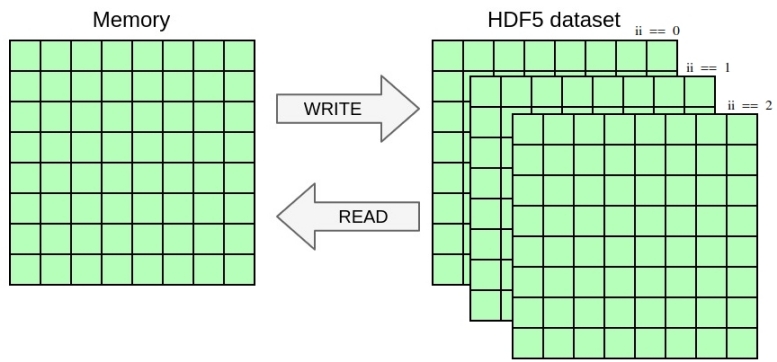
Running the current code in parallel should already work and yield one file per process containing the local data block. In this exercise you will write one single file with parallel HDF5 whose content should be independent from the number of processes used.
The mpi plugin was loaded to make sharing MPI communicators possible.
There are several tasks in this exercise:
communicator directive of the Dec'HDF5 plugin to switch to parallel I/O.main_field in datasets tree to take the global matrix into account. Hint: use psize.pcoord.Try to match the output from ex9.out, that should be independent from the number of processes used.
Touch only the yaml file in this exercise.
Here is graphical representation of the parallel I/O:
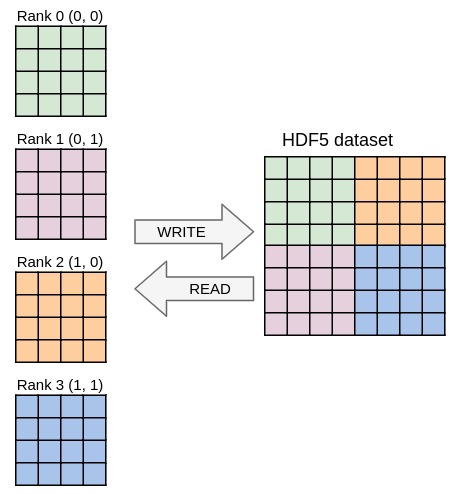
You can experiment with other plugins. Have a look at the PDI examples.